Using MatLab's Parallel Server for multi-core and multi-node jobs
MatLab users on SeaWulf can take advantage of multiple cores on a single node or even multiple nodes to parallelize their jobs. This FAQ article will explain how to do so using a "Hello World" example.
Single-node parallelization
To parallelize MatLab processes across a single node, we will write a MatLab script that uses "parcluster" and "parpool" to setup a pool of workers. Here is an example "Hello World" code for illustration:
% specify timezone to prevent warnings about timezone ambiguity
setenv('TZ', 'America/New_York')
% open the local cluster profile for a single node job
p = parcluster('local');
% open the parallel pool, recording the time it takes
tic;
parpool(p, 28); % open the pool using 28 workers
fprintf('Opening the parallel pool took %g seconds.\n', toc)
% "single program multiple data"
spmd
fprintf('Worker %d says Hello World!\n', labindex)
end
delete(gcp); % close the parallel pool
exit
Since we will only be using a single node, we will use the "local" cluster profile and set the parpool size to be equal to the number of cores on the compute node we'll be running on.
Let's save this MatLab script as "helloworld.m" and then write a Slurm batch script to execute it:
#!/usr/bin/env bash #SBATCH -p short-28core #SBATCH -N 1 #SBATCH --ntasks-per-node=28 #SBATCH --job-name=matlab_helloworld #SBATCH --output=MatLabHello28.log # load a MatLab module module load matlab/2021a # create a variable for the basename of the script to be run BASE_MFILE_NAME=helloworld # execute code without a GUI cd $SLURM_SUBMIT_DIR matlab -nodisplay -r $BASE_MFILE_NAME
In the above batch script, we load a MatLab module, create a variable to specify the basename of Hello World script, and then execute the script with the "-nodisplay" flag to prevent the GUI from trying to load. We've also selected one of the 28-core partitions to match the pool of 28 workers specified in the MatLab script.
Next, let's save this file as "helloworld.slurm" and submit the job script to the Slurm scheduler with "sbatch helloworld.slurm."
The output will be found in the log file "MatLabHello28.log":
< M A T L A B (R) >
Copyright 1984-2021 The MathWorks, Inc.
R2021a Update 5 (9.10.0.1739362) 64-bit (glnxa64)
August 9, 2021
To get started, type doc.
For product information, visit www.mathworks.com.
Starting parallel pool (parpool) using the 'local' profile ...
Connected to the parallel pool (number of workers: 28).
Opening the parallel pool took 24.7213 seconds.
Lab 1:
Worker 1 says Hello World!
Lab 2:
Worker 2 says Hello World!
Lab 3:
Worker 3 says Hello World!
Lab 4:
Worker 4 says Hello World!
Lab 5:
Worker 5 says Hello World!
Lab 6:
Worker 6 says Hello World!
Lab 7:
Worker 7 says Hello World!
Lab 8:
Worker 8 says Hello World!
Lab 9:
Worker 9 says Hello World!
Lab 10:
Worker 10 says Hello World!
Lab 11:
Worker 11 says Hello World!
Lab 12:
Worker 12 says Hello World!
Lab 13:
Worker 13 says Hello World!
Lab 14:
Worker 14 says Hello World!
Lab 15:
Worker 15 says Hello World!
Lab 16:
Worker 16 says Hello World!
Lab 17:
Worker 17 says Hello World!
Lab 18:
Worker 18 says Hello World!
Lab 19:
Worker 19 says Hello World!
Lab 20:
Worker 20 says Hello World!
Lab 21:
Worker 21 says Hello World!
Lab 22:
Worker 22 says Hello World!
Lab 23:
Worker 23 says Hello World!
Lab 24:
Worker 24 says Hello World!
Lab 25:
Worker 25 says Hello World!
Lab 26:
Worker 26 says Hello World!
Lab 27:
Worker 27 says Hello World!
Lab 28:
Worker 28 says Hello World!
Parallel pool using the 'local' profile is shutting down.
We can see from the above that there is a "Hello World" statement from each of the 28 workers.
Multi-node parallelization
It's also possible to use multiple nodes to run MatLab code via MatLab Parallel Server's built-in implementation of MPI, but this process requires a bit of intial setup:
1. First we need to load the matlab module and run the matlab executable in GUI mode:
module load matlab/2021a matlab
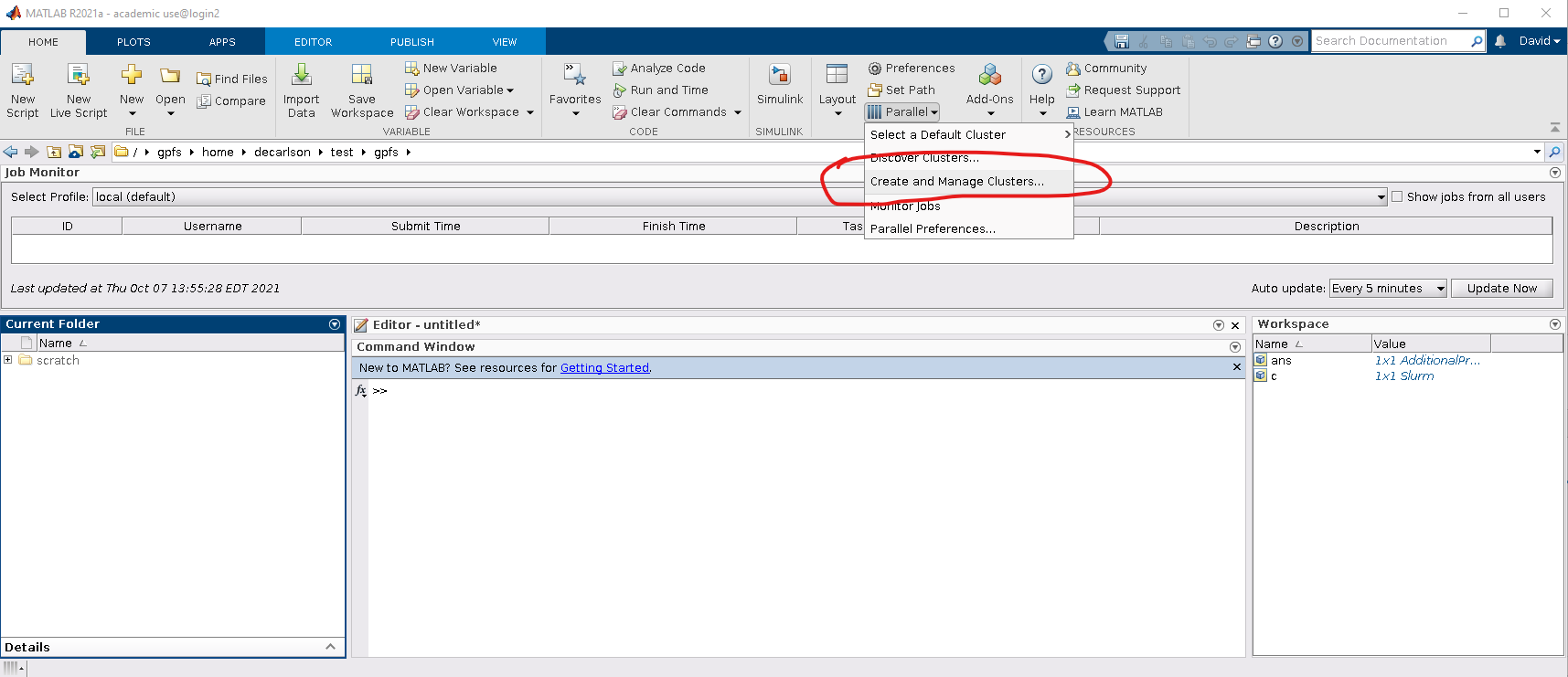
2. Next, within the matlab GUI, navigate to HOME->Parallel->Create and Manage Clusters:
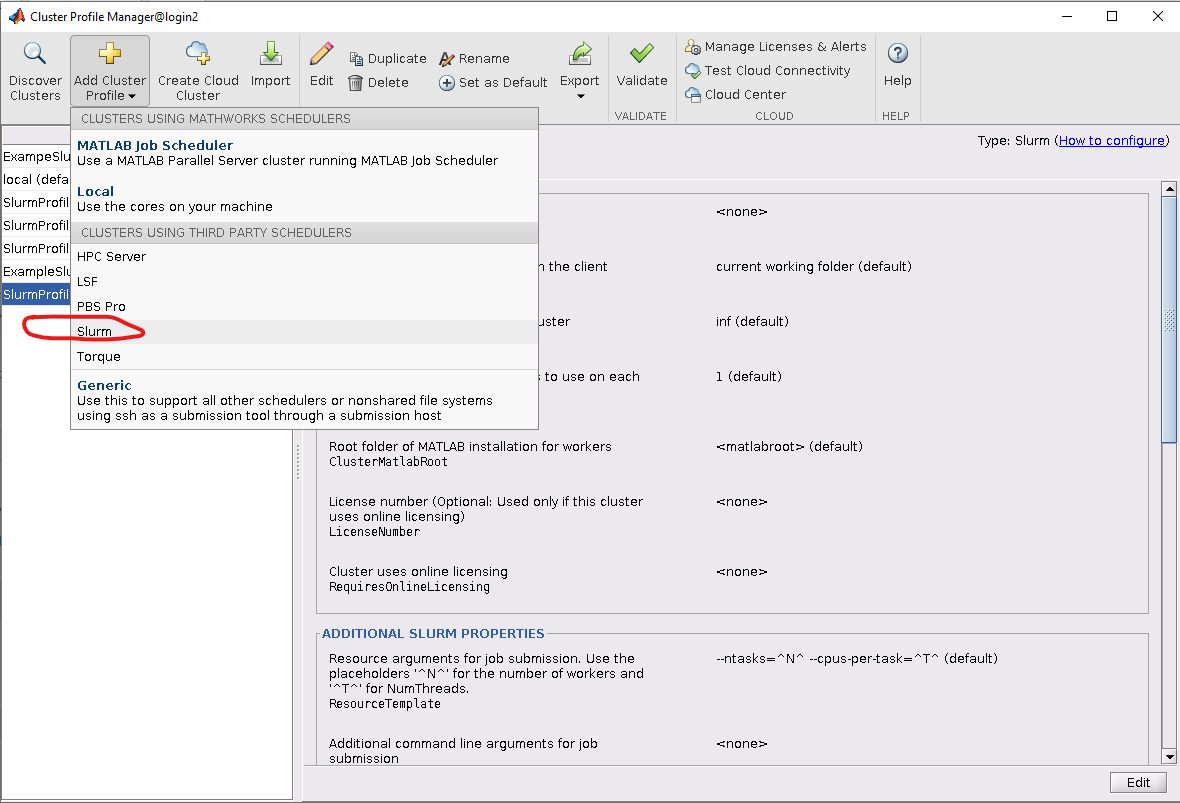
3. Next, we create a new cluster profile by navigating to "Add Cluster Profile" and choosing the Slurm scheduler:
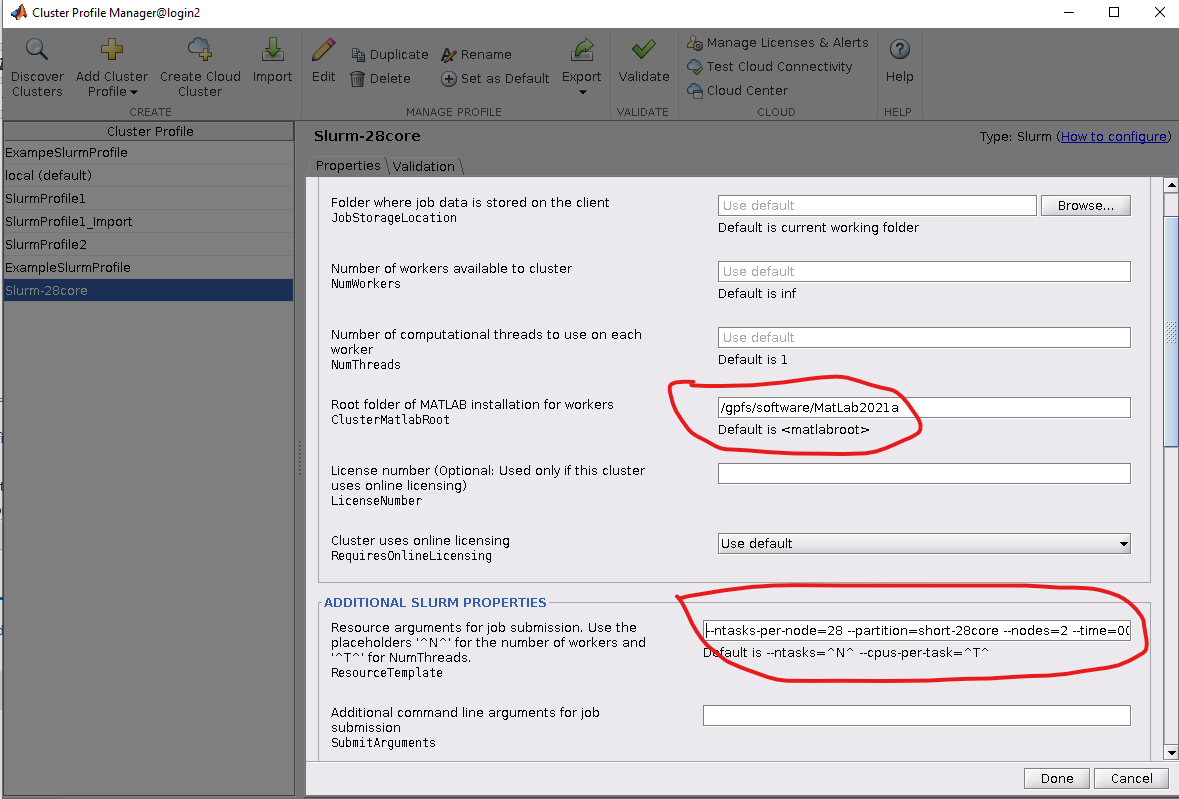
4. Finally, we edit the cluster profile to suite the needs of the job we will be running. Most of the settings can be left as default, though the following should be set:
matlabroot - the path to the MatLab root directory (e.g., /gpfs/software/MatLab2021a)
Resource arguments for job submission - these are the SBATCH flags used to control the job's behavior and resources utilized. At a minimum, the "--partition" flag should be set to a valid partition name.
An example Slurm profile that can be uploaded to SeaWulf, unzipped, and then imported within MatLab's Cluster Profile Manager window can be found here:
slurm-28core.mlsettings.zip
Finally, after editing the Slurm cluster profile to specify the resources needed for your job, we can launch the job with by executing a MatLab script at the command line (note that this can also be done within the MatLab GUI):
% specify timezone to prevent warnings about timezone ambiguity
setenv('TZ', 'America/New_York')
% open the desired Slurm cluster profile
p = parcluster('Slurm-28core');
% open the parallel pool, recording the time it takes
tic;
parpool(p, 56); % open the pool using 56 workers which will be spread across 2 nodes
fprintf('Opening the parallel pool took %g seconds.\n', toc)
% "single program multiple data"
spmd
fprintf('Worker %d says Hello World!\n', labindex)
end
delete(gcp); % close the parallel pool
exit
This time, we've specified the 'Slurm-28core' profile to the parcluster command. In addition, since the cluster profile is set to use 2 nodes in the short-28core partition, when we open the parallel pool we set the worker count to 56 workers.
Let's save this file as 'helloworld_slurm.m' and execute it:
matlab -nodisplay -r helloworld_slurm > helloworld_slurm.log
This will use MatLab's Slurm cluster profile to submit the code to the scheduler. The job may sit in the queue if the cluster is busy, but once it has run, we see a Hello World statement printed from 56 different workers printed in the log file:
... Lab 30: Worker 30 says Hello World! Lab 31: Worker 31 says Hello World! Lab 32: Worker 32 says Hello World! Lab 33: Worker 33 says Hello World! Lab 36: Worker 36 says Hello World! Lab 37: Worker 37 says Hello World! Lab 38: Worker 38 says Hello World! ...
Multi-node Slurm MatLab jobs can also be submitted using a specific Slurm MatLab plugin, which may offer more scriptable flexibility. To learn more about that option, please see here.

