RStudio Server provides a browser-based version of RStudio that lets you develop and run R scripts directly on SeaWulf’s compute nodes. It’s ideal for users who want to perform data analysis, visualization, or modeling using R without installing RStudio locally. You can access your home, scratch, and project directories and use the integrated terminal for command-line operations.
Getting Started
- Open the SeaWulf Open OnDemand portal: https://sn-ood.seawulf.stonybrook.edu/
- Log in using your NetID and DUO two-factor authentication.
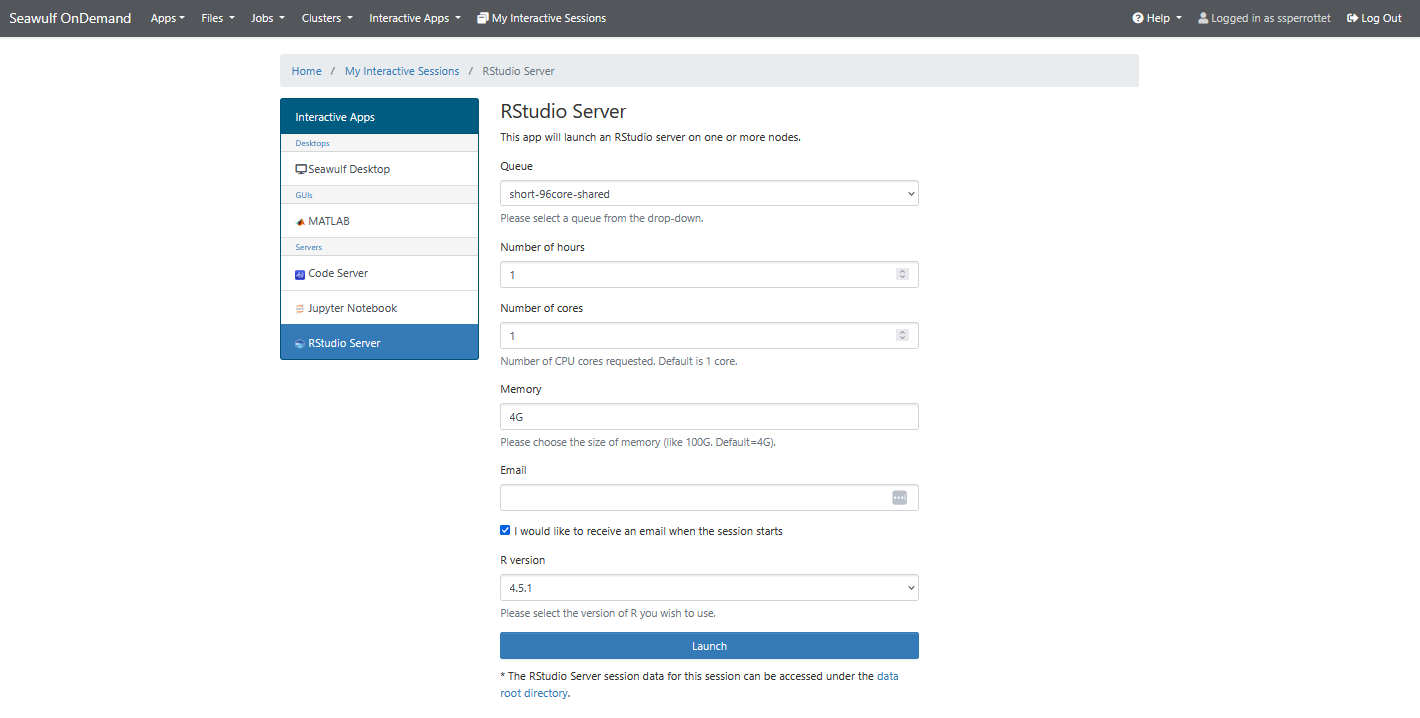
- Launch RStudio Server:
- Click the RStudio Server icon on the main dashboard
- OR navigate to Interactive Apps → RStudio Server
Session Configuration
- Queue: Choose a queue suited for your workload (e.g., short-40core-shared).
- Cores: 1–4 cores are typically enough for light to moderate work.
- Time Limit: Set the duration you expect to use the session.
- Memory: Adjust if your session involves large datasets or models.
- R Version: Select the R version you want to use.
- Email Notification: Optional; sends a message to your Stony Brook email when the session starts.
Tip: Request only the resources you need to reduce queue wait times.
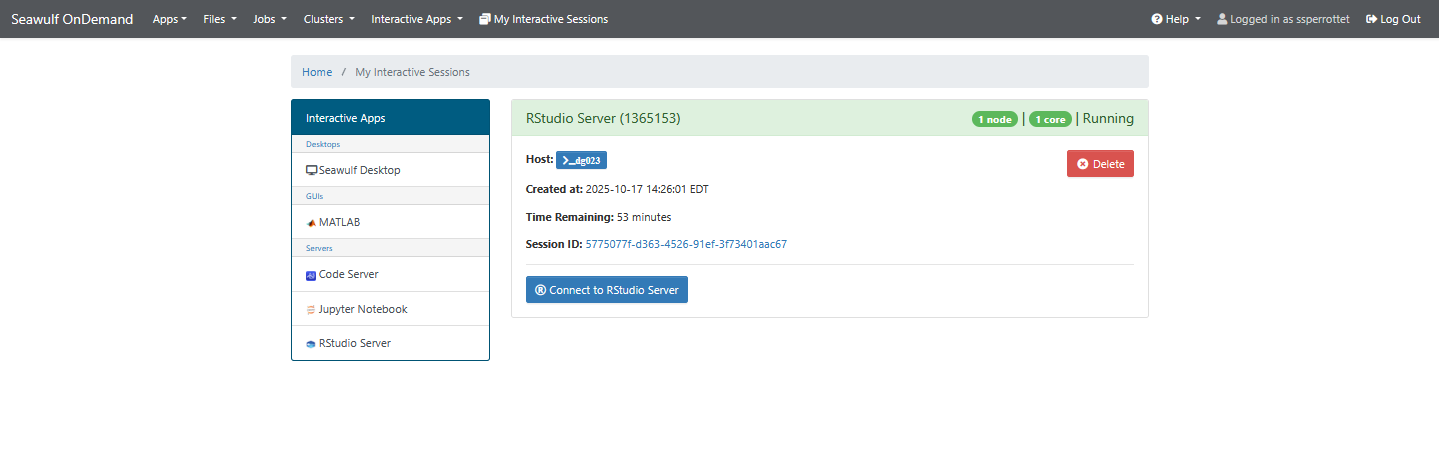
Click Launch to begin. Your session status will show Queued → Starting → Running. Once running, click Connect to RStudio to open the interface in a new browser tab.
Using RStudio Server
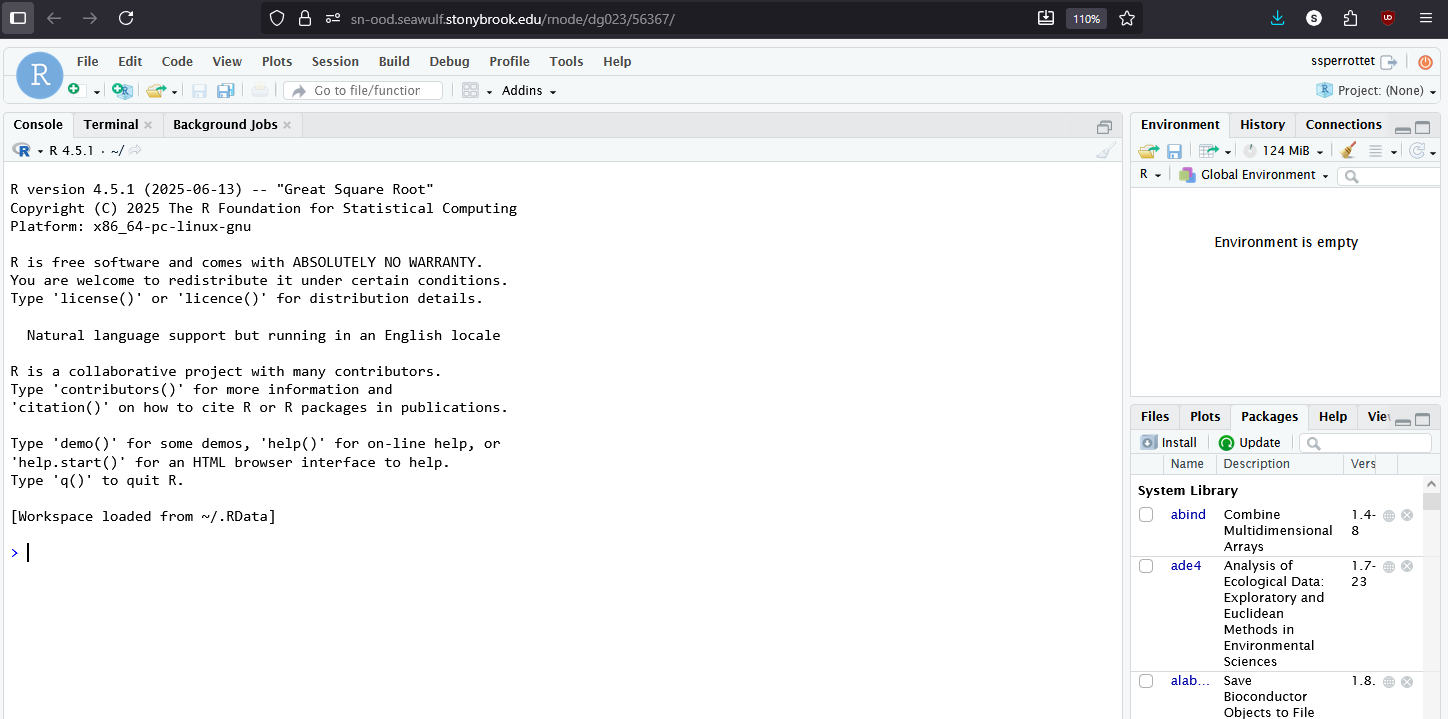
Once your session starts, you’ll have access to a full RStudio environment in your browser:
- Write, edit, and run R scripts in the editor pane.
- Use the console for interactive commands and quick calculations.
- Visualize data in plots and view results in the environment panel.
- Access your home, scratch, or project directories directly from the file browser.
- Use the integrated terminal for shell commands or job management.
Recommendations
- Save Work Frequently: Store scripts and results in your home or project directories to prevent data loss.
- Monitor Usage: Keep an eye on memory and CPU usage during heavy computations.
- Install Packages: Use
install.packages()to install libraries within your personal R environment. - Close Idle Sessions: End inactive sessions to free resources for other users.
Accessing Files & Collaboration
You can work directly with files in your home, scratch, or project directories. By default, only your home directory is visible. To access other spaces, create a symlink:
ln -s /gpfs/scratch/NetID /gpfs/home/NetID
Replace NetID with your actual NetID. This makes your scratch directory accessible in RStudio.
You can also use Git within RStudio to clone repositories, manage versions, and collaborate on code.
Note: Unlike MATLAB, RStudio sessions do not offer shareable links. Use Git or shared directories for collaboration.